EXERCISE 4
Exercise 4: Using mozbc and MOZART GCM data for initial and lateral boundary conditions for the HKH domain
***************************************************************************
***************************************************************************
- Save the wrf output data file from exercise 3 by moving it to a file with a different name.
mv wrfout_d01_2010-07-14_00:00:00 wrfout_d01_2010-07-14_save
- Obtain the mozbc utility as well as the MOZART model data.
cd /home/${USER}
cp -R /home/instructor1/CODE/MOZBC MOZBC
- Compile the mozbc utility. Take care to set your NETCDF_DIR either in your
environmental settings, or in the Makefile to the path to your netCDF library.
- Set the values in the the mozbc input data file. Take care to choose the correct chemistry for
the simulation. In this case, it is suggested you use the RADM2_SORGAM.inp file.
A common error when running mozbc is to not have all dates in the MOZART file. Confirm the dates in the
MOZART data file using the command:
ncdump -v date,datesec
It is recommend that one rename the MOZART netCDF data file from the NCAR/ACD download to remove the date and time stamp. A
simple name is prefered; something like h0010.nc
- Run mozbc to modify the wrfinput_d01 and wrfbdy_d01 data files. The program will modify the
files to provide the initial state and lateral boundary chemistry data using the MOZART GCM model output.
If you get an error message when running mozbc, it could be due to some of the chemical species being requested are not included
in the MOZART data file. For example, OH could be requested by the program, but not in the MOZART data. This would be due to
OH being a short lived species and best computed during the model run. So IF (big IF as it may not happen) one gets an error
regarding OH, then just remove OH from the RADM2_SORGAM.inp file speciation list and reruns the mozbc program. The same
would work for other short-lived species as well.
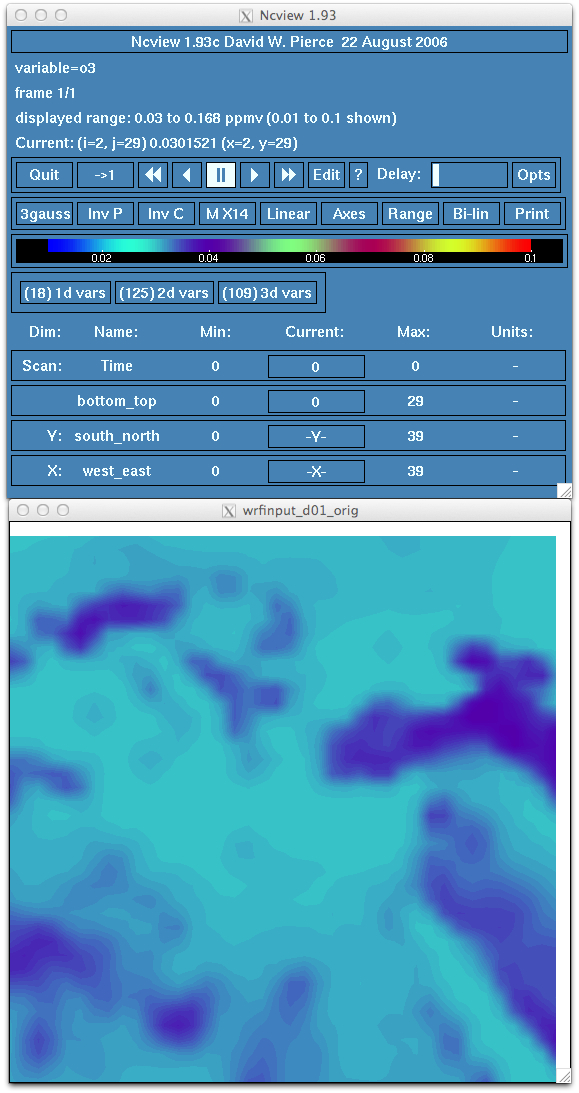
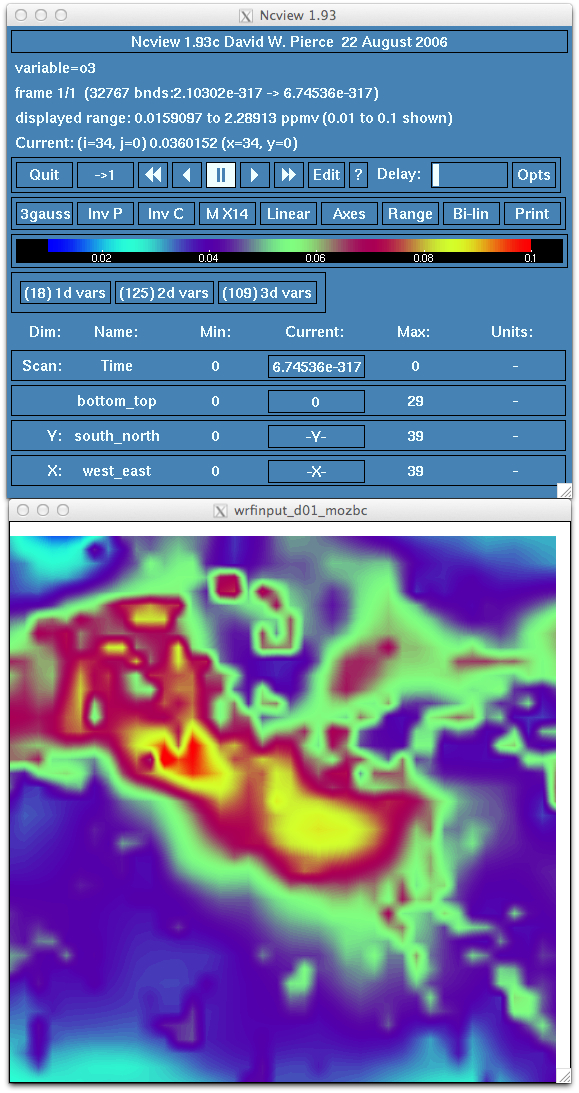
- Verify that the new intial chemical state and lateral boundary conditions has been changed and includes the MOZART data. For
example, look at surface ozone. Do you see an increase in ozone concentration? Does the range relfect a more realistic atmospheric
profile?
- Run wrf.exe using the new intial chemical state and lateral boundary conditions. Examine the model results and compare the wrfout file with the
wrfout file saved earlier in the exercise.
This concludes WRF-Chem emissions tutorial exercise 4
To compare solutions, download desired files from here.
Return to Nepal WRF-Chem Tutorial Page

