EXERCISE 3
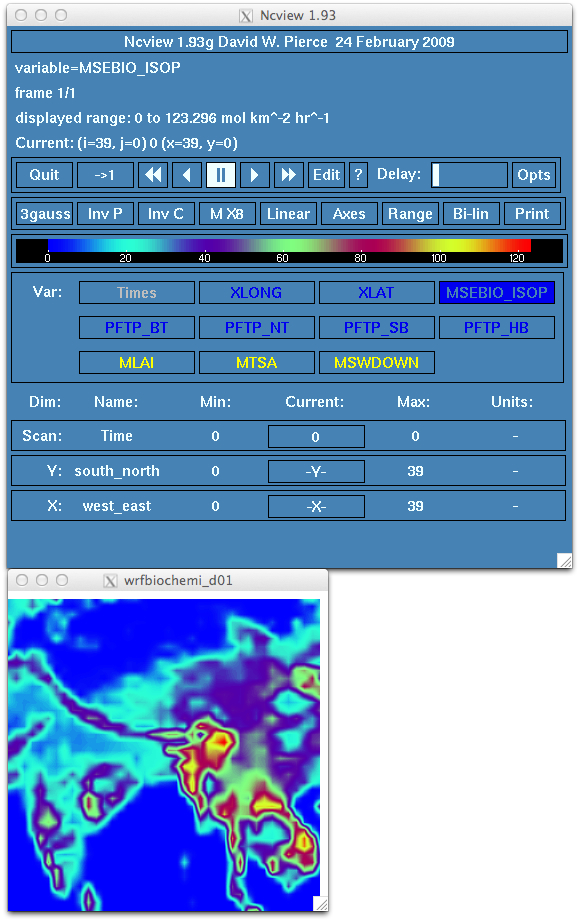
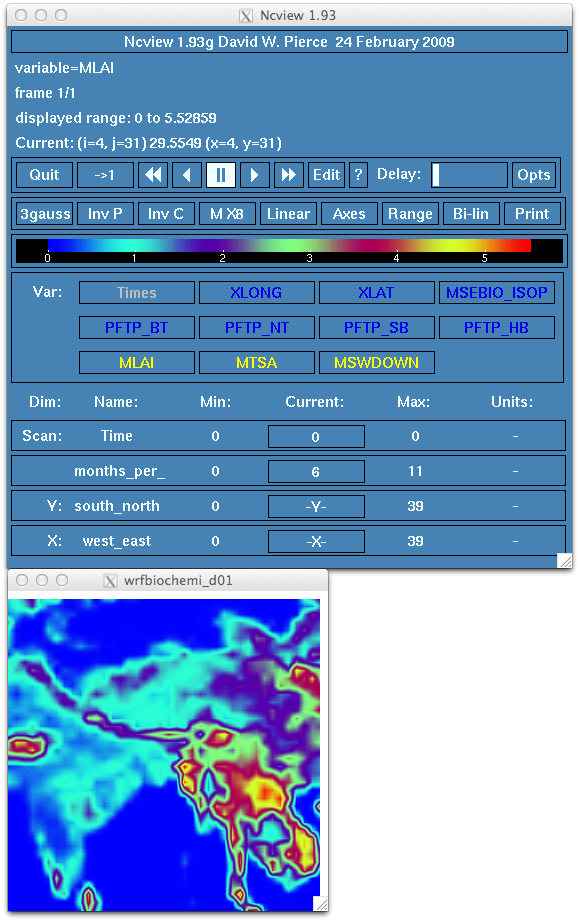
Exercise 3: Generating MEGAN biogenics to use along with the other emissions for the HKH domain.
***************************************************************************
mkdir MEGAN
cd MEGAN
tar -xf /home/instructor1/CODE/MEGAN_CODE/megan_tutorial.tar.gz
cd MEGAN/megan_code
Some compilers have a limit of 132 columns in the free formatted code. If you get an error message indicating an issue at column 132, then the code needs to be edited.
bio_emiss.f90:1231.132:
bal, attr_name(:slen), attr_xtype, attr_len, attrs(m)%attr_byte ), message
__________________________________________________________1
Error: Syntax error in argument list at (1)
To fix the problem, reduce the total columns in the code by splitting the code line in two and adding a continuating mark to the end of the first line. In this case, line 1231 of bio_emiss.f90 is too long and needs to be split into two lines. And of course, since the code was modified, you will need to clean and recompile the program.
For the tutorial, you should have something similar to:
&control
domains = 1,
start_lai_mnth = 1,
end_lai_mnth = 12,
wrf_dir = '/home/${USER}/WRFV3/test/em_real',
megan_dir = '/atmosdata/tempgroupdata/inputDATA/megan_data'
/
where one needs to use the actual user name in the file in place of ${USER}.
./megan_bio_emiss < megan_bio_emiss.inp >& run.out
Examine the run.out log file for any error messages. You should see the following message at the end of a successful run.
Finished megan2 bio emiss dataset shr200121_30sec.nc
map_megan2_emissions: Before write_bioemiss
map_megan2_emissions: After write_bioemiss
map_megan2_emissions: 3 cached grid(s)
(No, there is not a nice ending statement at this time.)


auxinput_5 -> Anthropogenic emissions
auxinput_6 -> Biogenic emissions (BEIS, MEGAN)
auxinput_7 -> Surface biomass burning fields
auxinput_8 -> GOCART background fields
auxinput_12 -> Chemistry initial fields
auxinput_13 -> Volcanic ash emissions
auxinput_14 -> Aircraft emissions
auxinput_15 -> Green House Gas emissions
This concludes WRF-Chem emissions tutorial exercise 3.